
My research at Indiana University in the laboratory of Dr. Armin Moczek has ultimately been motivated by one question: where does biodiversity come from? Usually, we answer this question by talking about the appearance, selection for, and fixation of random mutations over countless generations of individuals within a population. But what is an individual anyways? Rapidly accumulating evidence demonstrates the critical role microbial symbionts play in directing host development, and ultimately, fitness. This has generated the hypothesis that an individual is not a single organism, but rather the collective whole of host and associated microbes. But many questions remain: how applicable is this hypothesis across the tree of life? How does this new framework change our understanding of selection and evolution? And just how do we go about empirically testing this hypothesis? These are the types of questions that most excite me and have served to motivate my research.

Onthophagus beetles and the pedestal
My current study system, the Onthophagus dung beetle, is one uniquely suited to address my questions of interest. Despite generally feeding on nutritionally limited ruminant dung throughout all stages of their life, Onthophagus dung beetles have expanded onto every continent except Antarctica, specialized onto an enormous array of dung types, and speciated into well over 2000 extant species. A natural next question to ask given this constellation of facts is how this genus diversified so greatly, despite the inherent challenge of a dung-based diet. However, the early hypothesis that these beetles access their diet through association with microbial symbionts, has recently gained traction through experimental validation.
Work in O. taurus and the closely related genus Euoniticellus has demonstrated that the gut microbial communities of mothers and their offspring are highly similar, but distinct from the dung they feed on and the soil they live in; and that these gut microbes are reliably passed from mother to offspring through a pedestal – a fecal secretion onto which mothers oviposit their eggs. Shortly after hatching, larvae consume the pedestal before continuing on to feed upon the remainder of the brood ball provisioned for them by their mother. Parallel work has further demonstrated that the microbes found within the pedestal are functionally significant, as 1) Onthophagus beetles reared without their pedestal microbiota are slower to develop and eclose to smaller adults compared to individuals given access to their pedestals as larvae; 2) the negative growth consequences of pedestal-free development can be erased by re-inoculating larvae with pedestal derived bacteria cultivated in the laboratory.
To expand on this work, I sought to examine the extent to which different Onthophagus beetle species associate with distinct microbial communities, as might be expected if the microbiota has played a role in the diversification of these animals. My 2018 paper supported this prediction by showing that the exchange of the maternally provisioned microbiota between species resulted in negative developmental and survival outcomes, a fitness-relevant subset of which persisted into the next generation of unmanipulated animals.
The microbial community of Onthophagus beetles
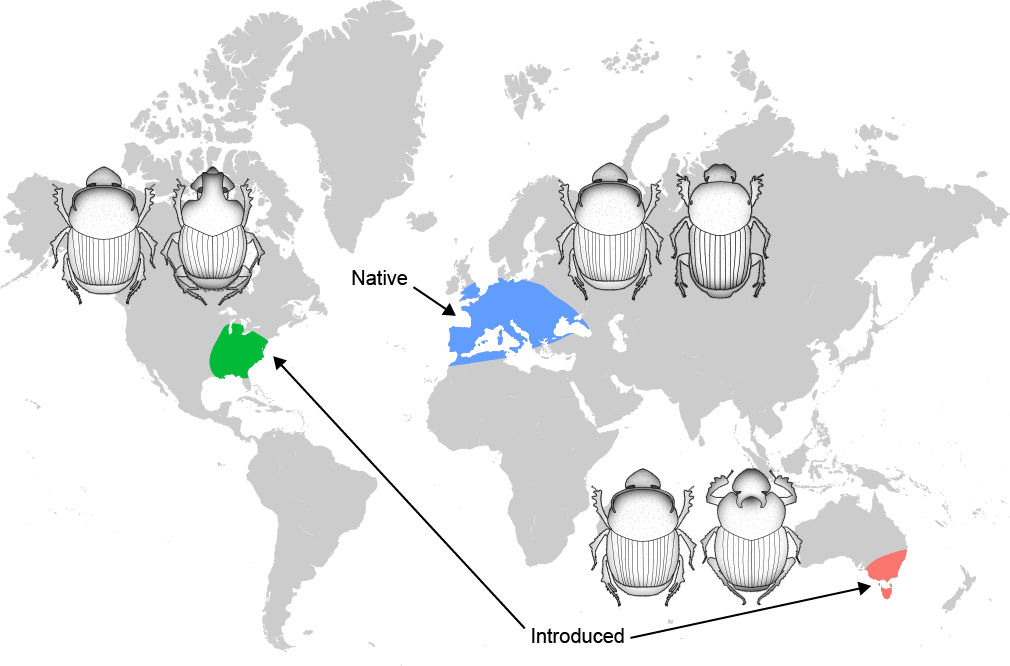
At this point, we knew that the pedestal was critical to Onthophagus development, and had hints that different species were coadapted to specific pedestal-transmitted microbiomes. But, we had virtually no information about the taxonomic identity of the microbial communities associated with different Onthophagus species and populations. To both address this shortcoming, while at the same time investigating if and how introduction events alter Onthophagus gut microbiota composition, I used 16S amplicon sequencing to compare gut microbe community profiles of O. taurus dung beetles collected from both their native and introduced ranges, alongside co-occurring native species. This project demonstrated that both evolutionary history and local environmental forces structure the microbial communities of these animals, but that the relative importance of these factos shaped by the specific circumstances that characterize individual introduction events. Furthermore, we identified microbial taxa such as Dysgonomonas that may constitute members of the core Onthophagus microbiome regardless of host population or species, but also Wolbachia which associates with Onthophagus beetles in a species or even population-specific manner. Beyond answering some fundamental questions about the structure of the Onthophagus microbiome, this project opened a number of new avenues to explore in this system - particularly the potential phenotypic effects of Wolbachia infection in this genus.
The role of the microbiota in rapid host adaptation, and speciation
The next chapters of my research are currently being written.
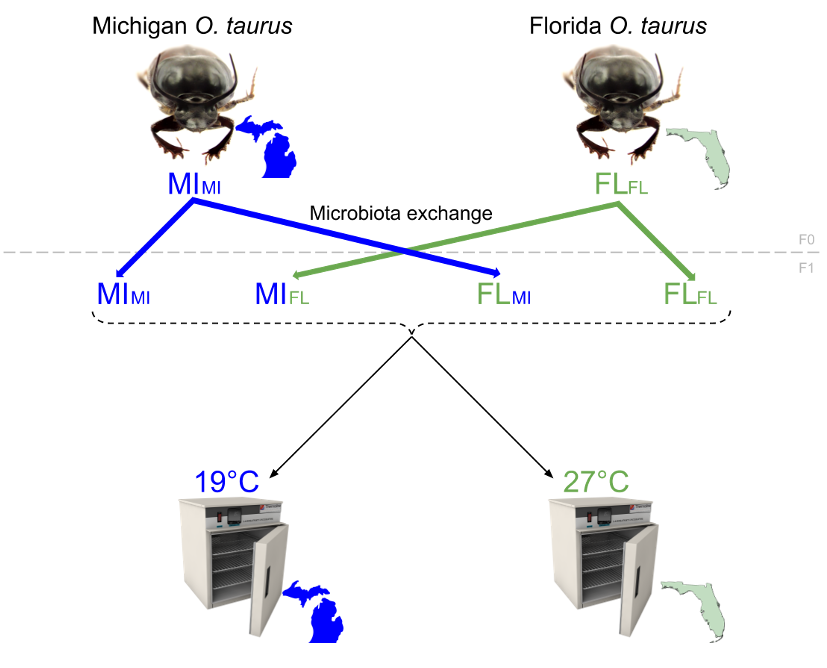
First, I am leveraging populations of Onthophagus taurus which in just 50 years have spread throughout the Eastern US from an accidental introduction site in Northern Florida to The Canadian border. This introduction event was unique among the many recorded for O. taurus in that it was not planned, and thus the animals came over with their full complement of pedestal-transmitted microbes. Furthermore, this is also the only introduction event where we have seen this species spread significantly outside its ancestral niche space. By coupling proven microbiome-manipulation techniques with ecologically relevant animal rearing, I’m aiming to uncover the role of the microbiota in the rapid environmental adaptation of O. taurus.
Second, I’m currently pursuing a project designed to tackle a question that has long intrigued me: do host-microbiota conflicts drive speciation by inducing hybrid lethality? While this area is of great interest to many biologists, empirical support is currently limited. I am addressing this shortcoming by assessing the impact of pedestal microbiota removal, and exchange between two members of an interbreeding but reproductively isolated Onthophagus species complex.
Both projects are nearing completion, and I am excited to share their results soon!

